Basic Data Processing with NumPy and Matplotlib¶
Overview, Objectives, and Key Terms¶
In this lesson, we’ll explore some core features of NumPy and Matplotlib
and extend our Python based “calculator” to include processing of
arrays of data. NumPy is the basic
numerical package for Python. A lot of its utility comes from its
numerical, multidimensional array type, ndarray. Use of these arrays
in a vectorized way often leads to significantly faster (and much more
compact) code. Moreover, the array-valued results of numerical
computations are easily visualized using Matplotlib, which was
originally developed to provide MATLAB-like graphics in the Python
environment.
I think it’s so important that students can create, process, and display data that I highlight it early in the course and revisit it throughtout. NumPy and Matplotlib together provide about the easiest way to work with data.
Objectives¶
By the end of this lesson, you should be able to
- define and manipulate one-dimensional NumPy arrays
- produce plots of data following best practices
- load data from text files
- save data to text files
Key Terms¶
numpyndarraynp.arraynp.onesnp.zerosnp.linspacenp.sinnp.mean(orv.meanwherevis of typendarray)max(ornp.maxorv.max)sum(ornp.sumorv.sum)matplotlib.pyplotnp.loadtxtnp.savetxtv.T, wherevis a two-dimensionalndarraystrformats
In [1]:
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
NumPy and One-Dimensional Arrays¶
NumPy is not part of Python, but it is a well-supported package. It comes by default in the Anaconda distribution, so you should already have it. Like all packages, we can import NumPy via
In [2]:
import numpy
However, it is far more common to use
In [3]:
import numpy as np
The difference between these approaches is in how we use the numpy
module. Built in to numpy are a number of same functions we saw in
math, e.g., sin. Hence, we could use numpy.sin(1.0), but if
we import numpy as np, we would use np.sin(1.0). I recommend
the latter because (1) it is shorter and (2) most online documentation
uses the np abbreviation.
Note: Useimport numpy as npinstead ofimport numpy.
A Motivating Example¶
Here’s a common task easily solved using NumPy: evaluate a mathematical
function f(x) at discrete points. Why discrete data? Perhaps because
that’s all the input data we have, e.g., from measurements, or perhaps
we’re trying to plot that function. In any case, this is where NumPy
excels. Consider the specific case for which we want to evaluate
\(f(x) = \sin(x)\) for \(x \in [0, 1]\), where x is to
limited to 10 evenly-spaced points.
Here’s what we need to do. First, always make sure NumPy is imported; in this notebook, we did that above, but here it is again for completeness.
In [4]:
import numpy as np
Now, we create an array x with 10, evenly-spaced points from 0 to 1
using the function np.linspace as follows:
In [5]:
x = np.linspace(0, 1, 10)
x # remember, a variable all by itself as input prints its value as output
Out[5]:
array([0. , 0.11111111, 0.22222222, 0.33333333, 0.44444444,
0.55555556, 0.66666667, 0.77777778, 0.88888889, 1. ])
Finally, we evaluate the function np.sin at these points.
In [6]:
y = np.sin(x)
y
Out[6]:
array([0. , 0.11088263, 0.22039774, 0.3271947 , 0.42995636,
0.52741539, 0.6183698 , 0.70169788, 0.77637192, 0.84147098])
Note that np.sin(x) is used and not math.sin(x). That’s because
x in this case is a ndarray, to which the base Python math
functions do not apply.
The function np.linspace(a, b, n) gives n evenly-spaced points
starting with a and ending with b. Writing the equivalent C++ or
Fortran to define such an x is a royal pain; that’s why Python+NumPy
(or MATLAB/Octave) is so nice for this type of problem.
Creating and Manipulating 1-D Arrays¶
We already saw one way to create a one-dimensional array, i.e.,
np.linspace. There are many other ways to create these arrays.
Suppose we have a list of numbers, e.g., 1.5, 2.7, and 3.1, and we’d
like to make an array filled with these numbers. That’s easy:
In [7]:
a = np.array([1.5, 2.7, 3.1])
a
Out[7]:
array([1.5, 2.7, 3.1])
(It turns out that [1.5, 2.7, 3.1] is actually a Python list,
but we’ll cover those later on when we need them; for now, we’ll stick
with NumPy arrays exclusively.)
The common ways to make 1-D arrays are listed in the table below.
| function | example use | what it creates creates |
|---|---|---|
np.array(seq
uence) |
np.array([1.5, 2.7, 3.1]) |
``array([ 1.5, 2.7, 3.1]) `` |
np.linspace(
start, stop, n
um) |
np.linspace(0, 1, 3) |
``array([ 0. , 0.5, 1. ]) `` |
np.ones(shap
e) |
np.ones(3) |
array([ 1., 1., 1.]) |
np.zeros(sha
pe) |
np.zeros(3) |
array([ 0., 0., 0.]) |
np.arange(st
art, end, step
) |
np.arange(1, 7, 2) |
array([1, 3, 5]) |
The table does not tell the whole story of these functions; use
help() or the ? mark for further details.
A useful aspect of NumPy arrays is that the basic arithmetic operations
of Python (think + and *) apply to such arrays via vectorized
operations. For example, two arrays of equal length can easily be
multiplied:
In [8]:
a = np.array([1, 2, 3])
b = np.array([2, 3, 4])
c = a*b
c
Out[8]:
array([ 2, 6, 12])
Individual arrays can also be operated on by arithmetic operaters. For
instance, we could double each element of a by doing
In [9]:
d = 2*a
d
Out[9]:
array([2, 4, 6])
Inspecting Arrays and Their Elements¶
What if we need to get the value of an array element? For example,
suppose, as done above, we doubled the array a, called it d, and
wanted the first value of d. We need to use array indexing via
the operator []. Here’s how we get that first value of d:
In [10]:
d[0]
Out[10]:
2
Yes–the first element is accessed using a zero. Python, like C, C++, and many other languages, is based on zero indexing, in which the numbering of sequences starts with zero. Get used to it!
Warning: Numbering starts with zero in Python
By using indexing, we can change the value of d[0], too, e.g.,
In [11]:
d[0] = 99
d
Out[11]:
array([99, 4, 6])
The number of elements in an array can be found in two ways. First, we
can use the built-in len function:
In [12]:
len(d)
Out[12]:
3
We can also use the size attribute, associated with any array,
that can be accessed through the . operator:
In [13]:
d.size
Out[13]:
3
If we want to access the last element of d, we can use that length
directly via
In [14]:
d[len(d)-1]
Out[14]:
6
but a much cleaner way to do so is via
In [15]:
d[-1]
Out[15]:
6
In fact, by using a negative number, the elements are accessed in
reverse starting from the last element. In other words, d[-1] is the
same as d[len(d)-1], d[-2] is the same as d[len(d)-2], and
so on.
Note: Access the \(i\)th to last element ofarrayviaarray[-(1+i)].
What else do arrays have to help us? Try using dir!
In [16]:
dir(d)
Out[16]:
['T',
'__abs__',
'__add__',
'__and__',
'__array__',
'__array_finalize__',
'__array_interface__',
'__array_prepare__',
'__array_priority__',
'__array_struct__',
'__array_ufunc__',
'__array_wrap__',
'__bool__',
'__class__',
'__complex__',
'__contains__',
'__copy__',
'__deepcopy__',
'__delattr__',
'__delitem__',
'__dir__',
'__divmod__',
'__doc__',
'__eq__',
'__float__',
'__floordiv__',
'__format__',
'__ge__',
'__getattribute__',
'__getitem__',
'__gt__',
'__hash__',
'__iadd__',
'__iand__',
'__ifloordiv__',
'__ilshift__',
'__imatmul__',
'__imod__',
'__imul__',
'__index__',
'__init__',
'__init_subclass__',
'__int__',
'__invert__',
'__ior__',
'__ipow__',
'__irshift__',
'__isub__',
'__iter__',
'__itruediv__',
'__ixor__',
'__le__',
'__len__',
'__lshift__',
'__lt__',
'__matmul__',
'__mod__',
'__mul__',
'__ne__',
'__neg__',
'__new__',
'__or__',
'__pos__',
'__pow__',
'__radd__',
'__rand__',
'__rdivmod__',
'__reduce__',
'__reduce_ex__',
'__repr__',
'__rfloordiv__',
'__rlshift__',
'__rmatmul__',
'__rmod__',
'__rmul__',
'__ror__',
'__rpow__',
'__rrshift__',
'__rshift__',
'__rsub__',
'__rtruediv__',
'__rxor__',
'__setattr__',
'__setitem__',
'__setstate__',
'__sizeof__',
'__str__',
'__sub__',
'__subclasshook__',
'__truediv__',
'__xor__',
'all',
'any',
'argmax',
'argmin',
'argpartition',
'argsort',
'astype',
'base',
'byteswap',
'choose',
'clip',
'compress',
'conj',
'conjugate',
'copy',
'ctypes',
'cumprod',
'cumsum',
'data',
'diagonal',
'dot',
'dtype',
'dump',
'dumps',
'fill',
'flags',
'flat',
'flatten',
'getfield',
'imag',
'item',
'itemset',
'itemsize',
'max',
'mean',
'min',
'nbytes',
'ndim',
'newbyteorder',
'nonzero',
'partition',
'prod',
'ptp',
'put',
'ravel',
'real',
'repeat',
'reshape',
'resize',
'round',
'searchsorted',
'setfield',
'setflags',
'shape',
'size',
'sort',
'squeeze',
'std',
'strides',
'sum',
'swapaxes',
'take',
'tobytes',
'tofile',
'tolist',
'tostring',
'trace',
'transpose',
'var',
'view']
Several names worth pointing out are dot, max, mean,
shape, and sum. The max, mean, and sum functions
have pretty intuitive names (and purposes):
In [17]:
d.max()
Out[17]:
99
In [18]:
d.mean() # here, (99+4+6)/3
Out[18]:
36.333333333333336
In [19]:
d.sum()
Out[19]:
109
It turns out that Python has max and sum as built-in functions.
Hence, we could instead do
In [20]:
max(d)
Out[20]:
99
In [21]:
sum(d)
Out[21]:
109
but not mean(d) because mean is not a built-in function.
However, mean is a NumPy function, so we could do
In [22]:
np.mean(d)
Out[22]:
36.333333333333336
There is no real difference between d.mean() and np.mean(d). My
own personal preference is the latter because it looks closer to the
programming I first learned eons ago.
The shape attribute is somewhat special; here it is for d:
In [23]:
d.shape
Out[23]:
(3,)
Here, the shape indicates a purely one-dimensional array. We’ll see
below that shape is a bit more interesting for two-dimensional
arrays.
Finally, the dot function implements the dot
product, sometimes called
the scalar product, for two arrays. In mathematics, we typically refer
to one-dimensional arrays of numbers as vectors. For two vectors c
and d of length \(n\), the dot product is defined as
where \(c_i\) is the \(i\)th element of \(\mathbf{c}\). In NumPy, that operation is done via
In [24]:
c.dot(d)
Out[24]:
294
or
In [25]:
np.dot(c, d)
Out[25]:
294
Of course, knowing how the dot product is defined, we could also use
basic element-wise arithmetic coupled with the built-in sum
function, e.g.,
In [26]:
sum(c*d)
Out[26]:
294
Diving into pyplot¶
Recall our motivating example: defined x at evenly spaced points,
and evaluate np.sin at those points. We printed the points and the
function at those points, but it’s often useful to show it via a
graph. Matplotlib gives us the tools to do
just that. The most common way to import Matplotlib (and, specifically,
its pyplot “submodule”) is via
In [27]:
import matplotlib.pyplot as plt
In other words, plt is used as an abbreviation to the longer
matplotlib.pyplot.
Note: Just like NumPy is often shorted tonp,matplotlib.pyplotis often used asplt.
The most basic plot is very easy to make using plt.plot:
In [28]:
plt.plot(x, y) # x and y were defined above
Out[28]:
[<matplotlib.lines.Line2D at 0x7f8865b60dd8>]
To show the plot, we need to use plt.show:
In [29]:
plt.show()
Multiple Curves and Best Practices¶
What about multiple curves? Let’s define a second curved equal to
np.cos evaluated at x:
In [30]:
z = np.cos(x)
To plot both y and z is also easy, and here, I’ve added a couple
extra features that define the basic ingredients needed for a good plot:
In [31]:
plt.plot(x, y, 'k-o', x, z, 'r--x')
plt.legend(['sin(x)', 'cos(x)'], loc=0)
plt.xlabel('x')
plt.show()
Out[31]:
[<matplotlib.lines.Line2D at 0x7f8865ae8ba8>,
<matplotlib.lines.Line2D at 0x7f8865ae8e48>]
Out[31]:
<matplotlib.legend.Legend at 0x7f8865af38d0>
Out[31]:
Text(0.5,0,'x')
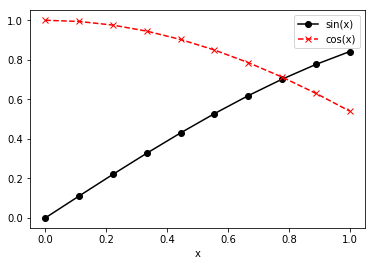
Let’s breakdown the new components. First, consider x, y, 'k-o'.
That means y is to be plotted against x, and the line to be used
is black (the k), a solid line (the -), and has circle markers
(the o). The second curve is defined by x, z, 'r--x'. Here, that
plots z against x with a red (the r), dashed (the --)
line with x’s as markers (the x). A legend is provided indicating
which line is which. The format of the legend always has a sequence of
str labels for each line enclosed in []‘s (again, that’s a
list in Python, but just follow the syntax for now. By setting
loc=0, we’re putting the legend in the best possible place to avoid
overlapping with the curves displayed. As a baseline rule, any plot
produced should contain an xlabel and either a ylabel (if a
single curve) or a legend (if multiple curves).
Note: All plots should include appropriate axis labels and legends.
Some of the available colors, line styles, and markers available in
plt.plot are summarized in the tables below.
| symbol | color |
|---|---|
k |
black |
b |
blue |
r |
red |
g |
green |
c |
cyan |
m |
magenta |
| symbol | style |
|---|---|
- |
solid |
-- |
dash-dash |
-. |
dash-dot |
: |
dot-dot |
| symbol | marker |
|---|---|
o |
circle |
x |
x |
^ |
triangle pointed up |
> |
triangle pointed right |
s |
square |
h |
hexagon |
In some applications, it can be very important to use a combination of colors, line styles, and markers that produce good looking and easily read graphics when viewed in color and in black and white formats.
Note: Choose good colors, line styles, and markers to ensure excellent contrast in all media.
Data Input and Output via NumPy¶
For many problems, the data we need to process lives in a file outside our Python code. There are a variety of ways to load data, but NumPy provides an easy way to load data that is in a relatively simple format. Consider a text file that has the following data:
time (s) vel (m/s) acc (m/s**2)
0.00000000 1.00000000 0.00000000
0.22222222 1.24884887 0.01097394
0.44444444 1.55962350 0.08779150
0.66666667 1.94773404 0.29629630
0.88888889 2.43242545 0.70233196
1.11111111 3.03773178 1.37174211
1.33333333 3.79366789 2.37037037
1.55555556 4.73771786 3.76406036
1.77777778 5.91669359 5.61865569
2.00000000 7.38905610 8.00000000
The structure is simple: three columns separated by white space. The first row contains information about what the data represent. Here, the columns correspond to times at which the velocity and acceleration of some object are given.
Loading Text Files¶
Go ahead and save this text in a file. Mine is called data.txt. To
load this file, we’ll use np.loadtxt. The basic use of this function
is just np.loadtxt(filename), where filename is a str that
contains the name of the file. Here, we need just a bit more. In
particular, we need tell NumPy to skip the very first row since it
contains regular text and not the numbers of interest. Here’s one way to
read in that text:
In [32]:
data = np.loadtxt('data.txt', skiprows=1)
data
Out[32]:
array([[0. , 1. , 0. ],
[0.22222222, 1.24884887, 0.01097394],
[0.44444444, 1.5596235 , 0.0877915 ],
[0.66666667, 1.94773404, 0.2962963 ],
[0.88888889, 2.43242545, 0.70233196],
[1.11111111, 3.03773178, 1.37174211],
[1.33333333, 3.79366789, 2.37037037],
[1.55555556, 4.73771786, 3.76406036],
[1.77777778, 5.91669359, 5.61865569],
[2. , 7.3890561 , 8. ]])
We have the data stored in the variable data. The type looks like an
ndarray, but the structure is more like a matrix, i.e., a
two-dimensional array—and it is! We can try out the shape
attribute again:
In [33]:
data.shape
Out[33]:
(10, 3)
That makes sense: we had 10 times at which the velocity and acceleration
were provided. 10 rows, and 3 columns. We’ll see next time how to access
entire rows and columns of multidimensional arrays, but there is a
better way to load in this data directly into three meaningful variables
t, v, and a.
In [34]:
t, v, a = np.loadtxt('data.txt', skiprows=1, unpack=True)
t
Out[34]:
array([0. , 0.22222222, 0.44444444, 0.66666667, 0.88888889,
1.11111111, 1.33333333, 1.55555556, 1.77777778, 2. ])
Now, we’ve got the times, velocities, and accelerations as three, separate, one-dimensional arrays. With NumPy, loading this sort of data is easy and recommended.
Note: If data is numerical and stored in a simple, collimated format, usenp.loadtxt
Like many functions in NumPy and other modules, np.loadtxt accepts
quite a large number of optional arguments. You need to use help or
the online documentation to learn more (and, of course, you’ll get to
explore these in the exercises.)
Saving Text Files¶
Saving data is just as easy as loading. The function of interest is
np.savetxt, and it requires some data and a file name. One way we
could save our t, v, and a arrays is via
In [35]:
np.savetxt('output_1.txt', [t, v, a])
Go and check output_1.txt. The data is there, for sure, but it’s not
quite the same format that we started with. Instead, t, v, and
a were written out in their own row. That’s fine, but often, we want
to spit out the data in pretty much the same way we’d like it to be read
into our programs.
One issue is that NumPy treats [t, v, a] like an array with a shape
of (3, 10), which is exactly how it’s saved to file. Such
two-dimensional arrays are basically matrices, and if we want the 3
and 10 reverse, we need the transpose. Here, we can first produce a
2-D array explicitly from [t, v, a] by using the np.array
function, i.e.,
In [36]:
data = np.array([t, v, a])
data
Out[36]:
array([[0. , 0.22222222, 0.44444444, 0.66666667, 0.88888889,
1.11111111, 1.33333333, 1.55555556, 1.77777778, 2. ],
[1. , 1.24884887, 1.5596235 , 1.94773404, 2.43242545,
3.03773178, 3.79366789, 4.73771786, 5.91669359, 7.3890561 ],
[0. , 0.01097394, 0.0877915 , 0.2962963 , 0.70233196,
1.37174211, 2.37037037, 3.76406036, 5.61865569, 8. ]])
Then, we need its transpose, i.e.,
In [37]:
data.T
Out[37]:
array([[0. , 1. , 0. ],
[0.22222222, 1.24884887, 0.01097394],
[0.44444444, 1.5596235 , 0.0877915 ],
[0.66666667, 1.94773404, 0.2962963 ],
[0.88888889, 2.43242545, 0.70233196],
[1.11111111, 3.03773178, 1.37174211],
[1.33333333, 3.79366789, 2.37037037],
[1.55555556, 4.73771786, 3.76406036],
[1.77777778, 5.91669359, 5.61865569],
[2. , 7.3890561 , 8. ]])
That appears to be what we want in our output (if we want it to look
like our original input). The only missing ingredient is the first row
that indicates what each column contains. Such information can be
included by using the header argument. By default, # is appended
to the beginning, since np.loadtxt actually ignores lines that start
with # (or some other user-specified comment symbol). Here, we can
avoid the comment symbol in our output by using setting the comment
argument to zero.
Here it all is together:
In [38]:
np.savetxt('output_2.txt', data.T, header='time (s) vel (m/s) acc (m/s**2)', comments='')
An Aside: String Formatting¶
The output is pretty darn close, but the numbers are longer (and they
were the first try, too). Behind the scenes, float values are stored
with about 15 digits of useful information. When we read in the values
from data.txt, they were defined only with 8 digits after the
decimal point. You can see that output_2.txt has pretty much the
same values, but there are some strange digits out beyond that eight
digit. We can limit the output to only 8 digits by using a string
format. Consider the following:
In [39]:
print("%.1f" % 0.1234)
print("%.4f" % 0.1234)
print("%.16f" % 0.1234)
print("%.3e" % 0.1234)
0.1
0.1234
0.1234000000000000
1.234e-01
Here, the basic syntact is a str format, which contains a %
followed by, e.g., .4f. That means to format the number in normal
decimal notation (and not, e.g., scientific) with 4 digits after the
decimal point. The last example used an e, which indicates
scientific notation (with, here, three digits after the decimal point
and before the e). String formatting can be very powerful (and
complex). We’ll turn again to the topic again later on, but this gives
you a taste!
Saving Text Files: Revisited¶
Back to our example. Armed with a bit of formatting, here’s the final solution for getting output identical to our original input:
In [40]:
np.savetxt('output_3.txt', data.T, header='time (s) vel (m/s) acc (m/s**2)', comments='', fmt='%.8f')
Inspection of output_3.txt confirms it’s exactly like our original
data.txt.
Further Reading¶
Make sure to explore in depth the help output of all the functions
explored in this lesson. Many times, not all function arguments need to
be used, but it’s very helpful to know that they exist and what they can
help you to do.